multishifter
casm powered utility to create gamma surfaces, UBER curves, and twisted bilayers along crystal planes.
Tutorial I: Slicing a structure
In this tutorial we’ll use multishift slice to create a structure of \(\mathrm{Mg}\) with ab-vectors in the \((0,\bar{1},1)\) plane (pyramidal slip plane).
Mg primitive cell
Current implementations of multishifter work exclusively with the VASP structure format.
Download the primitive cell for \(\mathrm{Mg}\) here, or create a file called mg.vasp with the following crystallographic data:
multishifter tutorial i
1.00000000
1.59609453 2.76451683 0.00000000
-1.59609453 2.76451683 0.00000000
0.00000000 0.00000000 5.18401957
Mg
2
Direct
0.6666667 0.6666667 0.7500000 Mg
0.3333333 0.3333333 0.2500000 Mg
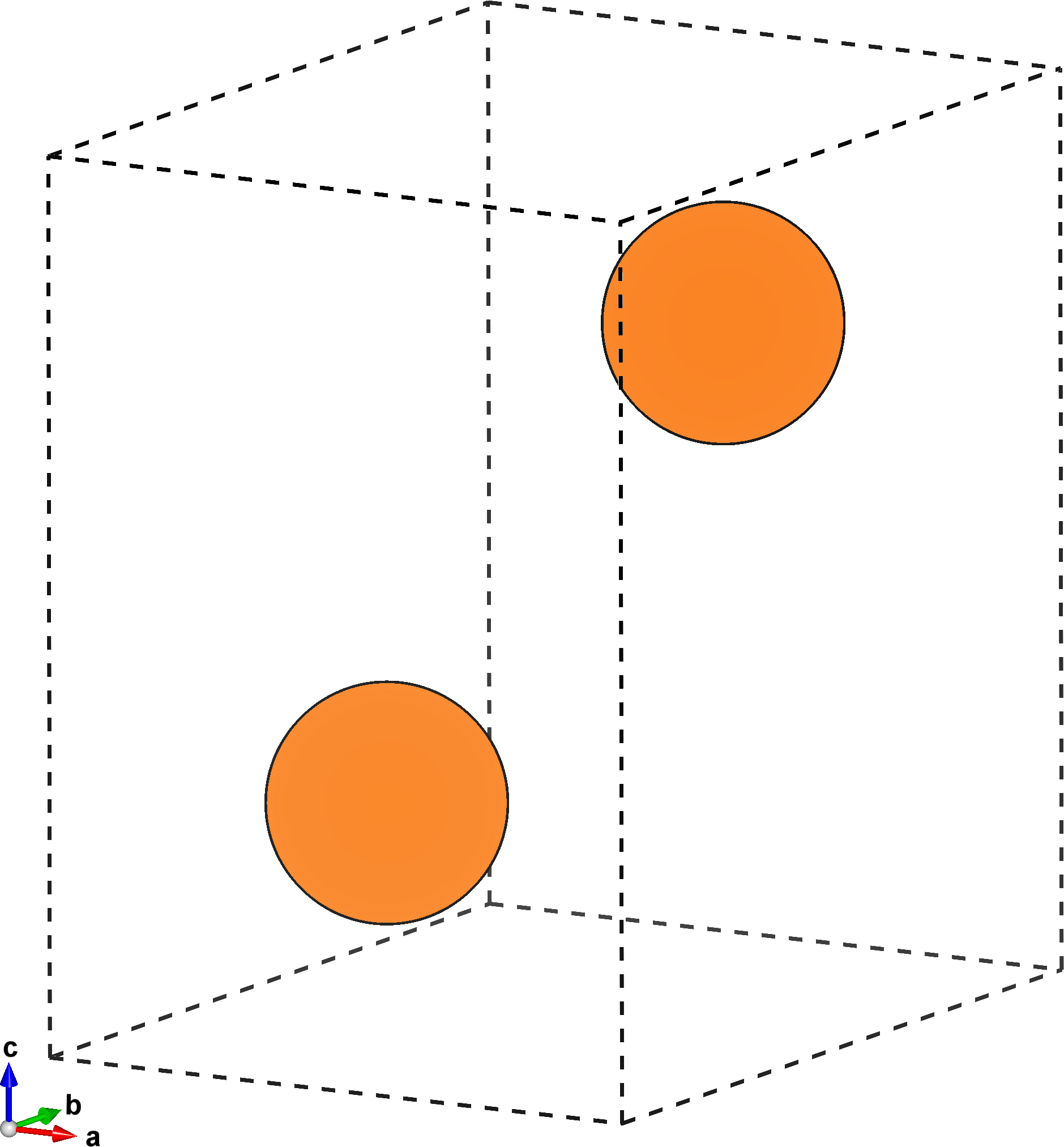
The figure below on the left shows the primitive unit cell of \(\mathrm{Mg}\), while the one on the right highlights the prismatic plane. This is the slip plane we’re interested in, and has miller indices \((0,\bar{1},1)\).
 |
Slice the structure
From your working directory, run
multishift slice --input mg.vasp --millers 0 -1 1 --output mg_pyramidal.vasp
This will read mg.vasp as the starting structure, and slice it allong the specified miller indexes, saving the generated structure to mg_pyramidal.vasp.
Output
You will now have a file named mg_pyramidal.vasp.
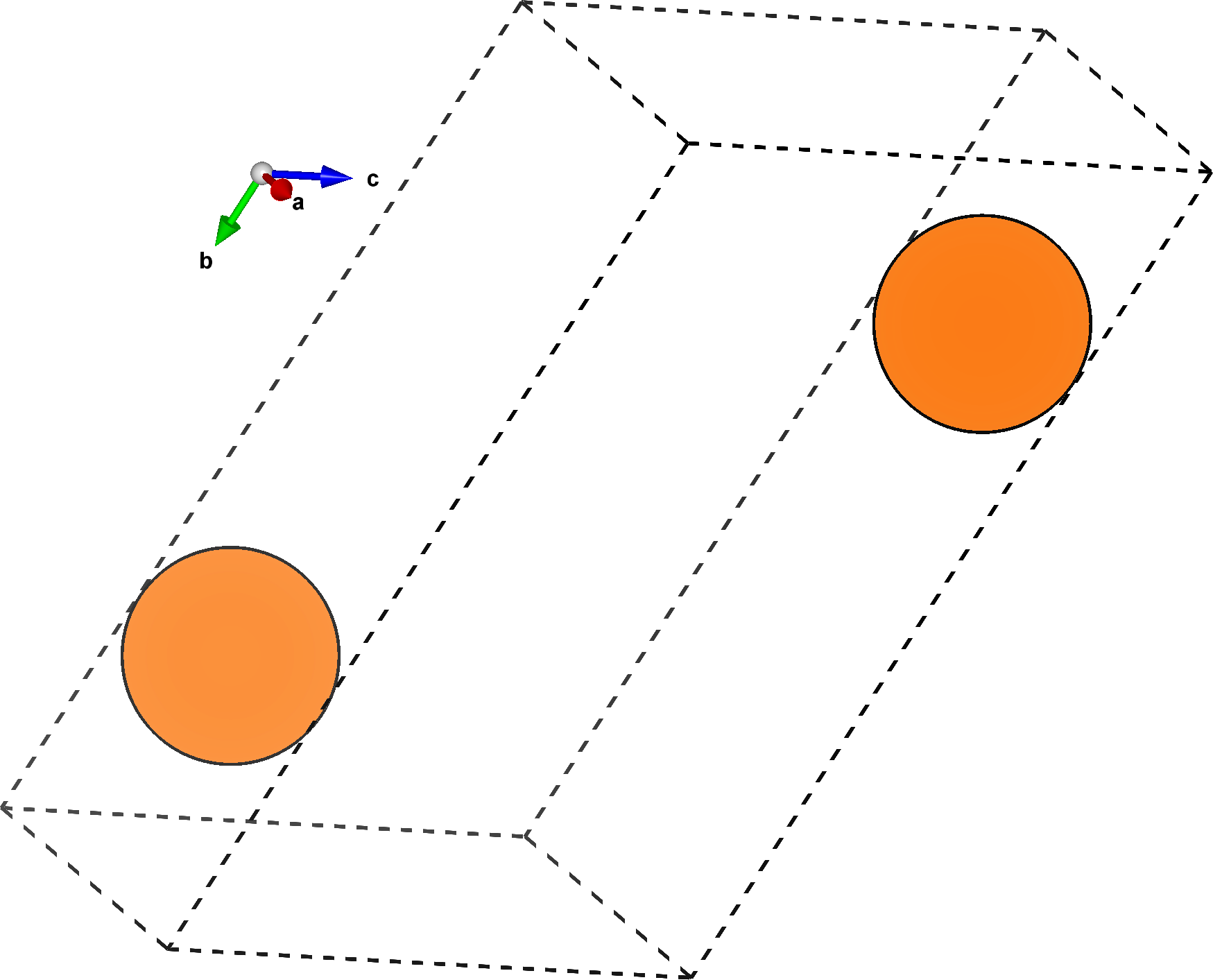
The plane defined by the millers argument is spanned by the \(ab\) vectors of this structure.
By default, the slice command also performs a rigid rotation on the final structure, aligning the \(a\) vector along the \(x\) direction, and the \(b\) vector in the \(xy\) plane.
This final rotation can be supressed by using the flag --dont-align.
The rotation can also be applied to any structure using multishift align.
 |
Slicing graphite
In the previous example, the sliced structure was still a primitive cell, albeit with an unconventional definition. If we slice the primitive cell of graphite along the \((1,1,1)\) layers, we’ll see this isn’t always the case.
Download the primitive cell for graphite here, or create a file called graphite.vasp with the following cyrstallographic data:
multishifter tutorial i.b
1.0
4.2654600143 0.0000000000 0.0000000000
3.5512614760 2.3627719025 0.0000000000
3.5512614760 1.0734450299 2.1048531614
C
2
Direct
0.166667989 0.166668006 0.166667997
0.833331925 0.833332067 0.833331971
You can repeat the slice command with new parameters now:
multishift slice --input graphite.vasp --millers 1 1 1 --output graphite_sliced.vasp
The resulting structure is a size 3 superstructure of the starting primitive structure, but has \(ab\) vectors parallel to the individual graphene layers. The figure below contrasts the primitive cell with the sliced structure.
