multishifter
casm powered utility to create gamma surfaces, UBER curves, and twisted bilayers along crystal planes.
Tutorial IV: Cleaving for UBER
In this tutorial we’ll use multishift cleave to generate many slab structures separated by varying amounts of empty space from each other.
Calculations of these structures can be used to determine how surface energy changes as a function of slab separation.
Al slab cell
We’ll start with a slab of \(\mathrm{Al}\) with 8 atomic layers.
The exposed surface is the \((1,1,1)\) plane of the conventional cell, which is just the \((0,0,1)\) direction of the primitive cell.
You can download it here, or create a file called al_stack8.vasp yourself with the follwing data:
Al stack
1.00000000
2.85595465 0.00000000 0.00000000
1.42797732 2.47332928 0.00000000
-0.00000000 1.64888618 18.65501764
Al
8
Direct
0.00000000 0.00000000 0.00000000 Al
0.37500000 0.25000000 0.12500000 Al
0.75000000 0.50000000 0.25000000 Al
0.12500000 0.75000000 0.37500000 Al
0.50000000 -0.00000000 0.50000000 Al
0.87500000 0.25000000 0.62500000 Al
0.25000000 0.50000000 0.75000000 Al
0.62500000 0.75000000 0.87500000 Al
You can also create your own slab from a primitive cell using multishift stack, as explained in a previous tutorial.
Cleave your slab
The cleave command takes a list of separation distances in \(\AA\) that will be inserted between the periodic images of the starting slab.
We’ll give 3 different values, and specify al_cleave as the directory where everything should be written to:
multishift cleave --input al_stack8.vasp --values -1.5 0.0 2.0 --output al_cleave
Once you’ve run this command, the output directory al_cleave will be created, which has the following structure:
al_cleave
├── slab.vasp
├── record.json
├── cleave__2.000000
│ └── POSCAR
├── cleave__-1.500000
│ └── POSCAR
└── cleave__0.000000
└── POSCAR
The first file slab.vasp is simply a backup of the input structure al_stack8.vasp.
The second file record.json holds a short description for each of the generated structures, and is described in detail in a separate tutorial.
The generated structures are each saved to an idividual directory named cleave__*, with the cleavage value fromatted into the directory name.
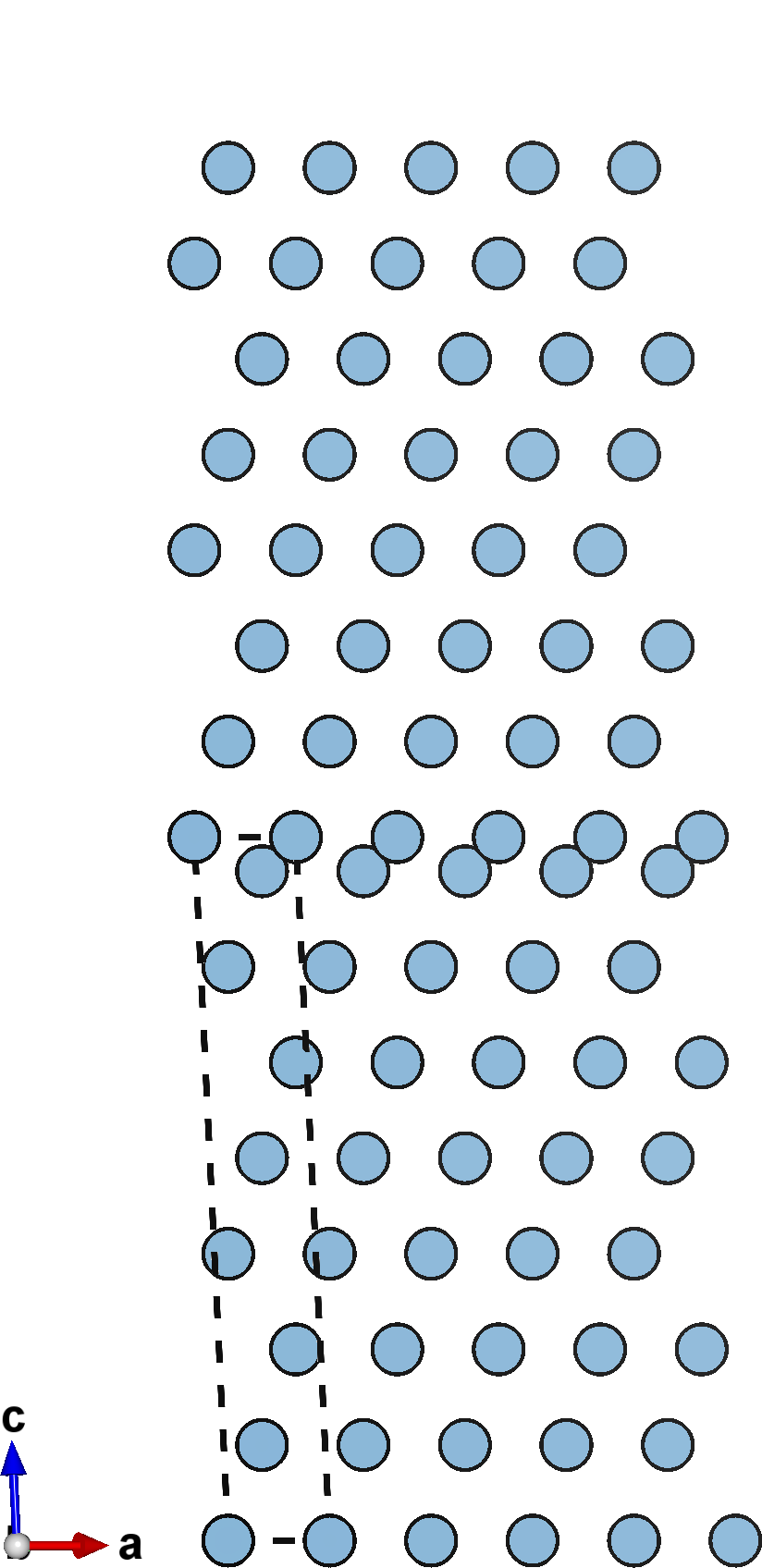
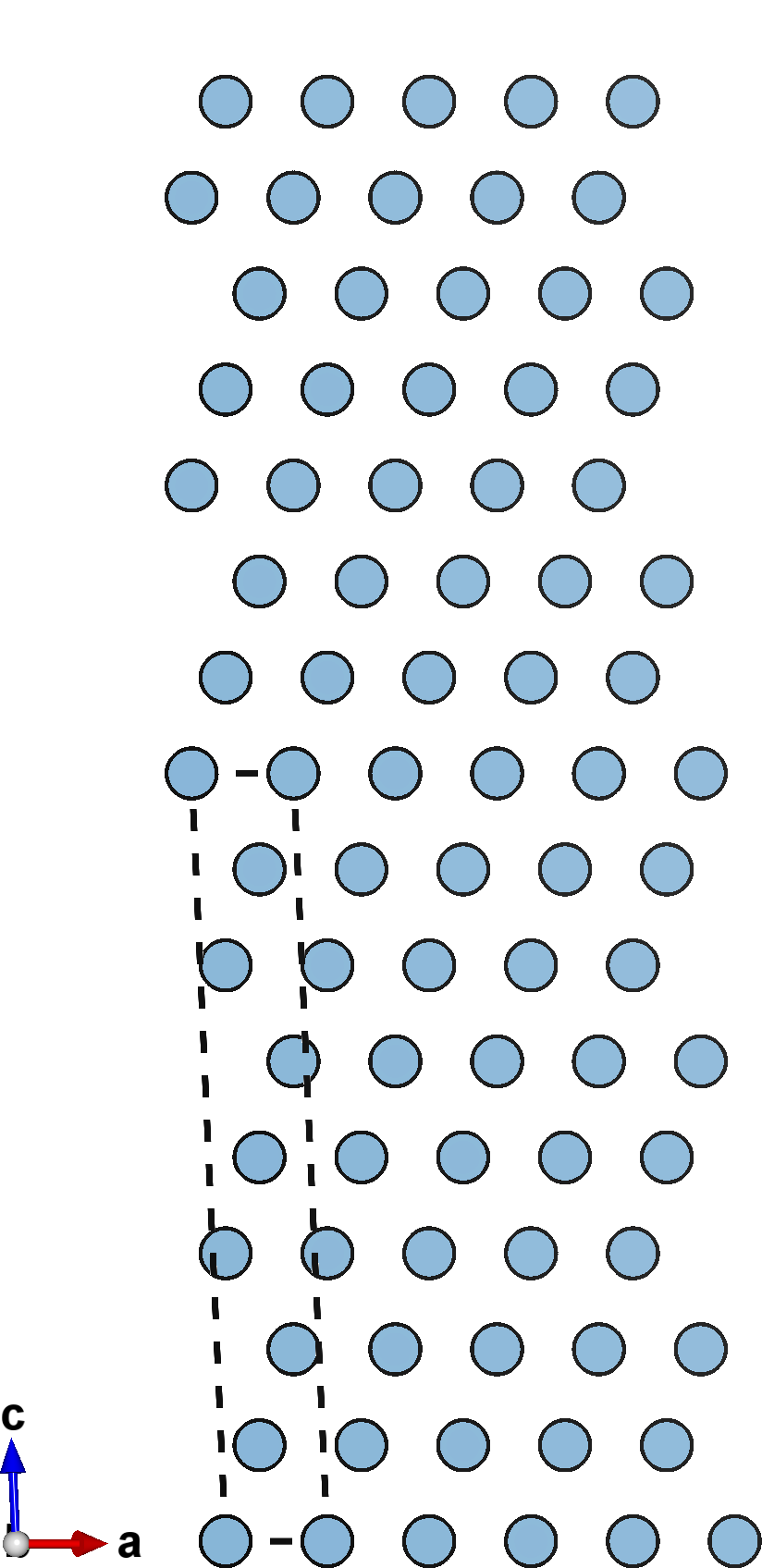
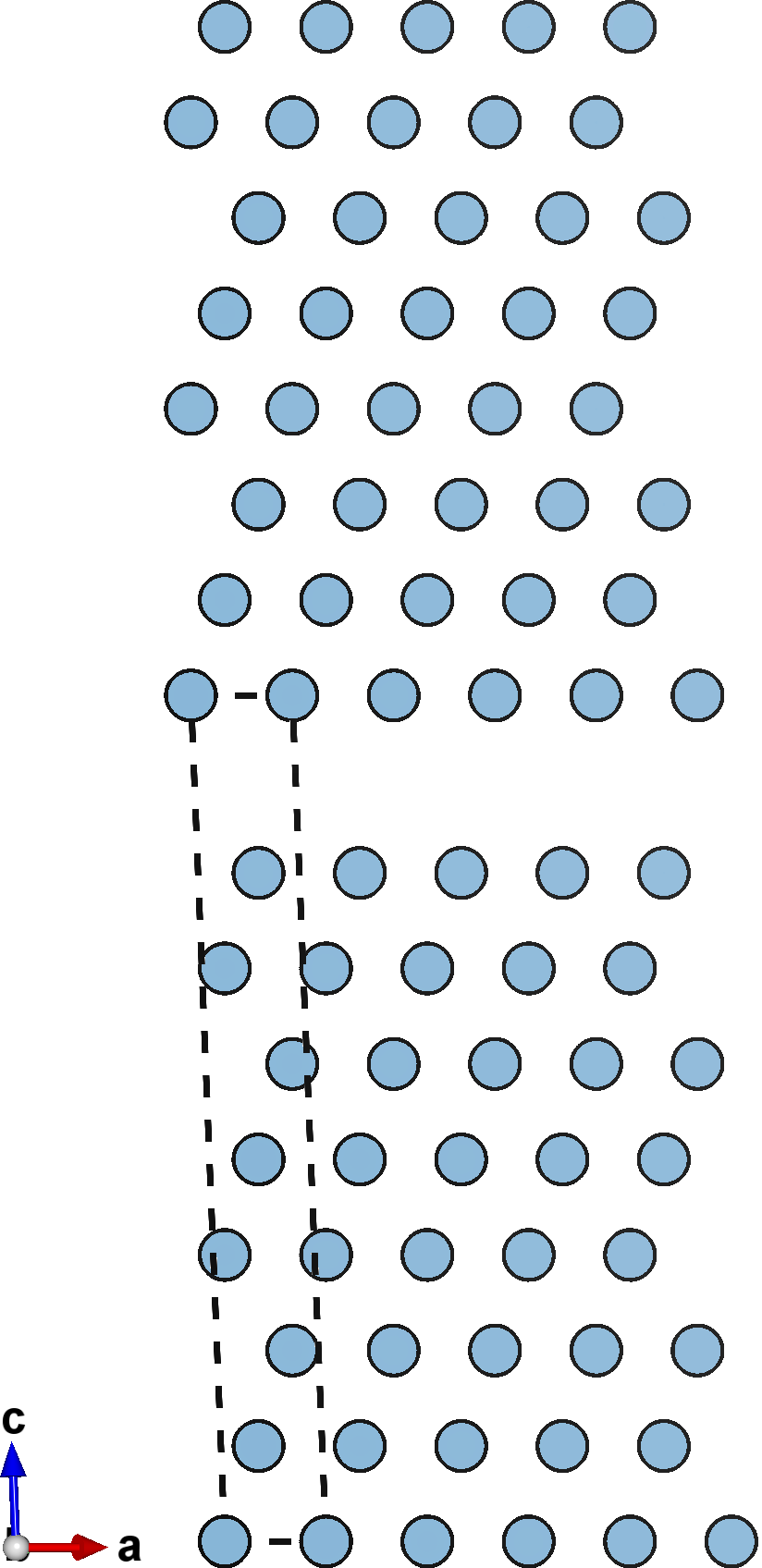
A negative cleavage value (left) causes the slabs to compress into each other, reducing the spacing between them. A null cleavage value (center) does nothing, and the output structure is simply an unperturbed copy of the input slab. A positive cleavage value (right) inserts space bewtween the slabs, separating them away from each other.
 |
 |
 |
A worked example
With enough cleavage values, you can fit an UBER curve. The following command generates 26 cleavage values, with smaller increments concentrated around the equilibrium distance.
rm -r al_cleave
multishift cleave --input al_stack8.vasp --output al_cleave --values -0.15 -0.1 -0.05 0.0 0.05 0.1 0.15 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0 1.25 1.5 1.75 2.0 2.5 3.0 3.5 4.0 6.0 8.0
The image below shows the energy values of each structure calculated with DFT, as a function of separation distance for the \(Al\) slab. As the cleavage value increases, the energy converges to twice the value of the surface energy of the material (two surfaces per slab). The data for these calculations and the script used for the plot are avaiable for you to download.
